
CAMEO - Duotang

CAMEO - Duotang
Duotang, a genomic epidemiology analyses and mathematical modelling notebook
Pillar 6 - CAMEO, CoVaRR-Net
29 September, 2023
Survey request
Doutang, VirusSeq Data portal, and Viral AI would like to improve your user experience. Please take a few minutes to respond to this survey.
SARS-CoV-2 In Canada
Introduction
This notebook was built to explore Canadian SARS-CoV-2 genomic and epidemiological data with the aim of investigating viral evolution and spread. It is developed by the CAMEO team (Computational Analysis, Modelling and Evolutionary Outcomes Group) associated with the Coronavirus Variants Rapid Response Network (CoVaRR-Net) for sharing with collaborators, including public health labs. These analyses are freely available and open source, enabling code reuse by public health authorities and other researchers for their own use.
Canadian genomic and epidemiological data will be regularly pulled from various public sources (see list below) to keep these analyses up-to-date. Only representations of aggregate data will be posted here.
Important limitations
These analyses represent only a snapshot of SARS-CoV-2 evolution in Canada. Only some infections are detected by PCR testing, only some of those are sent for whole-genome sequencing, and not all sequences are posted to public facing reposittories. Sequencing volumes and priorities have changed during the pandemic, and the sequencing strategy is typically a combination of prioritizing outbreaks, travellers, public health investigations, and random sampling for genomic surveillance.
For example, specific variants or populations might be preferentially sequenced at certain times in certain jurisdictions. When possible, these differences in sampling strategies are mentioned but they are not always known. With the arrival of the Omicron wave, many jurisdictions across Canada reached testing and sequencing capacity mid-late December 2021 and thus switched to targeted testing of priority groups (e.g., hospitalized patients, health care workers, and people in high-risk settings). Therefore, from this time onward, case counts are likely underestimated and the sequenced virus diversity is not necessarily representative of the virus circulating in the overall population.
Thus, interpretation of these plots and comparisons between health regions should be made with caution, considering that the data may not be fully representative. These analyses are subject to frequent change given new data and updated lineage designations.
The last sample collection date is 16 September, 2023
Current situation
There continues to be a substantial diversity of SARS-CoV-2 in Canada, with EG.5.1 subvariants, including EG.5.1.1 the most prevalent in many regions, along with XBB.1.16.6, EG.6.1, FL.1.5.1, and a mix of some some other XBB subvariants all with growing immune evasive mutations. The immune evasive S:F456L mutation is in the vast majority of samples (now over 75%) and continues to steadily increase in prevalence. The mutation S:478R is occurring in multiple variants showing growth. Multiple subvariants of XBB.1.16, XBB.1.9, XBB.1.5 and CH.1.1, have independently obtained the two mutations S:F456L and S:L455F (of interest due to their potential to provide additional immune evasion - and referred to as “FLIP” mutations due to their “exchange of L and F) and are now growing significantly in multiple regions, including the EG.5.1.1 subvariant HK.3 and the CH.1.1 subvariant DV.7.1. However, such collective variants with these”FLIP” mutations are still in low numbers (up to 15% of cases, except in NL and NB where they are up to 40%) so they will not be having an impact yet.
Sequences are also still being monitored closely to identify or track any highly divergent variants (variants with sudden, large mutational changes. This includes BA.2.86, tracking additional mutations of concern that are arising in it.
Variants of current interest, due to their current/potential growth advantage, mutations of potential functional significance, or spread in other countries (Bold only denotes changes since the last Duotang release - not necessarily those of greatest interest):
- DV.7.1 (CH.1.1 subvariant with S:F456L and S:L455F)
- EG.5.1 (XBB.1.9.2 with S:F456L plus S:Q52H plus some notable non-spike mutations)
- EG.5.1.1 (EG.5.1 subvariant that has been growing significantly versus other current variants).
- EG.5.1.3 (Another EG.5.1 subvariant growing notably)
- HK.3 (EG.5.1.1 with with S:F456L and S:L455F)
- JJ.1 (EG.5.1.4 with S:F456L and S:L455F - still being reported often as EG.5.1.4)
- FL.1.5.1 (XBB.1.9.1 with S:701V, ORF1a:G993S, S:456L and S:478R, with an interest in those with S:L455F) and other possible FL.1 subvariants with S:478R)
- XBB.1.16 which has S:T478R (with a particular interest in those with S:F456L like XBB.1.16.6 and also S:L455F)
- XBB.1.9.1, XBB.1.9.2, FL.5 (which have non-spike mutations of note, with a particular interest in those with S:Q613H OR with S:F456L plus NS6:Y49H (aka orf6:y49h)
…plus any highly divergent variants (like most recently BA.2.86) and sublineages with additional combinations of the mutations below.
- S:Q:52H (due to association with EG.5.1)
- S:L455F (in particular if present with S:F456L)
- S:F456L (evidence of increased immune evasion versus recent variants and a mutation growing in prevalence in multiple lineages in multiple regions)
- S:T478R (aka S:K478R - the S:T478K mutation occurred first). Evidence of increased infectivity when introduced into XBB.1.5. Associated with XBB.1.16 (which has additional mutations like S:E180V that may counteract this mutations advantage) but now seen in additional variants like XBB.2.3.
- S:P521S (evidence it could increase human ACE2 receptor binding/infectivity - associated with XBB.2.3 variants)
- S:Q613H (growing and seen in XBB.1.5, CH.1.1 and XBB.1.9.1)
- ORF1b:D1746Y (aka NSP14_D222Y - in XBB.1.16 variants)
- Orf1b:G662S aka NSP12_671 (in the context of a BA.2.86 subvariant) which has been found previously in other variant backgrounds to boosts viral replication at lower temperatures https://doi.org/10.1016/j.celrep.2023.113077
- ORF9b:I5T (note its a synonymous mutation in the overlapping N gene)
- ORF9b:N55S (synonymous mutation in N)
Plus other mutations identified through deep mutation scanning and the SARS-CoV-2 RBD antibody escape calculator. See:
- https://jbloomlab.github.io/SARS2-RBD-escape-calc/
- Greaney, Starr, & Bloom, Virus Evolution, 8:veac021 (2022)
- Cao et al, Nature, 614:521-529 (2023)
- Yisimayi et al, bioRxiv, DOI 10.1101/2023.05.01.538516 (2023)
With thanks to the global team of variant hunters which play a key role in identifying new variants of note.
Omicron sublineages in Canada
There are 368 unique named variants currently circulating in Canada since 2023-05-19 (last 120 days). Please see Pango lineage table for number of sequences per lineage present.
Here we take a look, sub-dividing the major sub-lineages currently circulating in Canada. (Click and drag to zoom, double click to reset. Clicking on an item in the legend will hide it, double clicking an item in legend will hide everything else but that item.)
Last 120 days
Last 120 days sublineages starting from 2023-05-19 (Frequency Table Download)
BA.1
BA.1 sublineages (Frequency Table Download)
BA.2
BA.2 sublineages (Frequency Table Download)
BA.4
BA.4 sublineages (Frequency Table Download)
BA.5
BA.5 sublineages (Frequency Table Download)
Recombinants
Recombinants sublineages (Frequency Table Download)
Selection on Omicron
Here we examine the relative rate of spread of the different sublineages of Omicron currently in Canada. Specifically, we determine if a new or emerging lineage has a selective advantage (s), and by how much, against a reference lineage (XBB.1.5) previously common in Canada (see the methods for more details about selection and how it is estimated).
Currently, the major group of Omicron lineages rising in frequency are the XBB.* group. This group was previously dominated by the XBB.1.5* lineages, which peaked in April 2023. A variety of other XBB* have been spreading since, and we now track the frequencies of XBB.1.5*, EG.5*, and XBB.Other, as well as all remaining non-XXB lineages. See Fastest Growing Lineages for details on lineages showing positive selection.
Left plot: y-axis is the proportion of these sub-lineages over time. Right plot: y-axis describes the logit function, log(freq(EG.5*, XBB.Other, or non-XBB*)/freq(XBB.1.5*)), which gives a straight line whose slope is the selection coefficient if selection is constant over time (see methods).
For comparison, Alpha had a selective advantage of s ~ 6%-11% per day over preexisting SARS-CoV-2 lineages, and Delta had a selective advantage of about 10% per day over Alpha.
Caveat: These selection analyses must be interpreted with caution due to the potential for non-representative sampling, lags in reporting, and spatial heterogeneity in prevalence of different sublineages across Canada. Provinces that do not have at least 20 sequences of a lineage during this time frame are not displayed.
Canada
Canada


BC
British Columbia


AB
Alberta


SK
Saskatchawan

MB
Manitoba


ON
Ontario


QC
Quebec


NS
Nova Scotia


NB
New Brunswick


NL
Newfoundland and Labrador


NULL
Case count trends by variant
These plots follow reported cases per 100,000 individuals (green dots), ignoring the most recent case counts (hollow circles), which are generally underestimated as data continues to be gathered. A cubic spline is fit to the log of these case counts to illustrate trends (top curve). The last two days of fitted case counts are then used to estimate the daily exponential growth rate \(r\) in COVID-19 cases. The fit from the “Selection on Omicron” section above is used to show how each of the sub-lineages is growing or shrinking, with the corresponding growth rate \(r\) for each sub-lineage on the last two days of fitted case counts. Note that only a small fraction of cases are currently being tested by PCR, so the y-axis height is underestimated by orders of magnitude (e.g., by 92-fold in BC mid-2022, Skowronski et al. 2022). Thus, graphs should only be used to describe growth trends and not absolute numbers. For detailed methodology, please see the methods section in the appendix.
Canada
Canada

BC
British Columbia

AB
Alberta

SK
Saskatchawan

MB
Manitoba

ON
Ontario

QC
Quebec

NS
Nova Scotia

NB
New Brunswick

NL
Newfoundland and Labrador

NULL
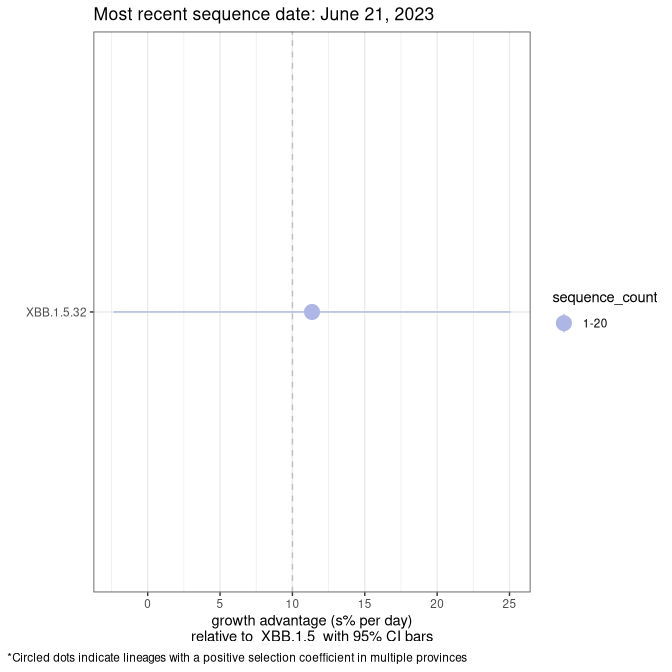
Fastest growing lineages
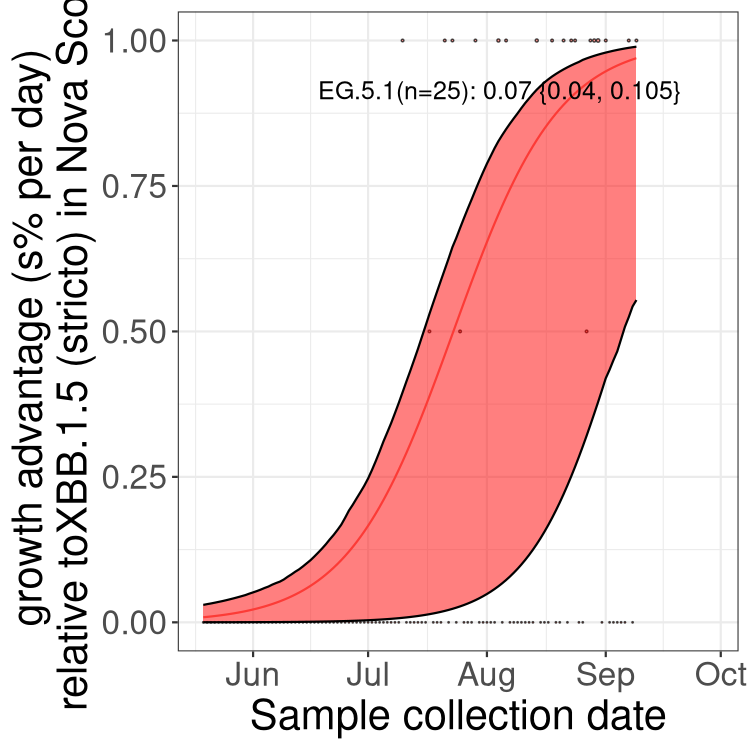
Here we show the selection estimates and their 95% confidence intervals for Pango lineages with more than 10 sequences in Canada since 2023-05-19 and with enough data to estimate the confidence interval. Each selection estimate measures the growth rate relative to XBB.1.5 stricto (i.e., sequences designated as XBB.1.5 and not its descendants, like XBB.1.5.1). Plots showing the change in variant frequency over time in Canada are given below for lineages with more than 50 sequences.
Plot (stricto)
Growth advantage of 0-5% corresponds to doubling times of more than two weeks, with 5-10% reflecting one to two week doubling times and over 10% representing significant growth of less than one week doubling time. Note that estimating selection of sub-variants with low sequence counts (less than 100 dots) is prone to error, such as mistaking one-time super spreader events or pulses of sequence data from one region as selection. Estimates with lower sequence counts in one region should be considered as very preliminary. For Canada-wide plot, a dot with a circle border indicates lineages with a positive selection coefficient in multiple provinces.
Canada
Plot single lineages in Canada *

BC
Plot single lineages in British Columbia

AB
Plot single lineages in Alberta

SK
Plot single lineages in Saskatchawan

MB
Plot single lineages in Manitoba
ON
Plot single lineages in Ontario

QC
Plot single lineages in Quebec

NS
Plot single lineages in Nova Scotia

NB
Plot single lineages in New Brunswick

NL
Plot single lineages in Newfoundland and Labrador

Plot (non stricto)
This plot highlights the groups of related lineages that are growing fastest (e.g., BQ.1* is the monophyletic clade that includes BQ.1.1 and all other BQ.1 sublineages.
Again, Growth advantage of 0-5% corresponds to doubling times of more than two weeks, with 5-10% reflecting one to two week doubling times and over 10% representing significant growth of less than one week doubling time. Note that estimating selection of sub-variants with low sequence counts (less than 100 dots) is prone to error, such as mistaking one-time super spreader events or pulses of sequence data from one region as selection. Estimates with lower sequence counts in one region should be considered as very preliminary. For Canada-wide plot, a dot with a circle border indicates lineages with a positive selection coefficient in multiple provinces.
Canada
Plot single lineages in Canada

BC
Plot single lineages in British Columbia

AB
Plot single lineages in Alberta

SK
Plot single lineages in Saskatchawan

MB
Plot single lineages in Manitoba

ON
Plot single lineages in Ontario

QC
Plot single lineages in Quebec

NS
Plot single lineages in Nova Scotia

NB
Plot single lineages in New Brunswick

NL
Plot single lineages in Newfoundland and Labrador

Table of all the selection estimates
Sublineages selection
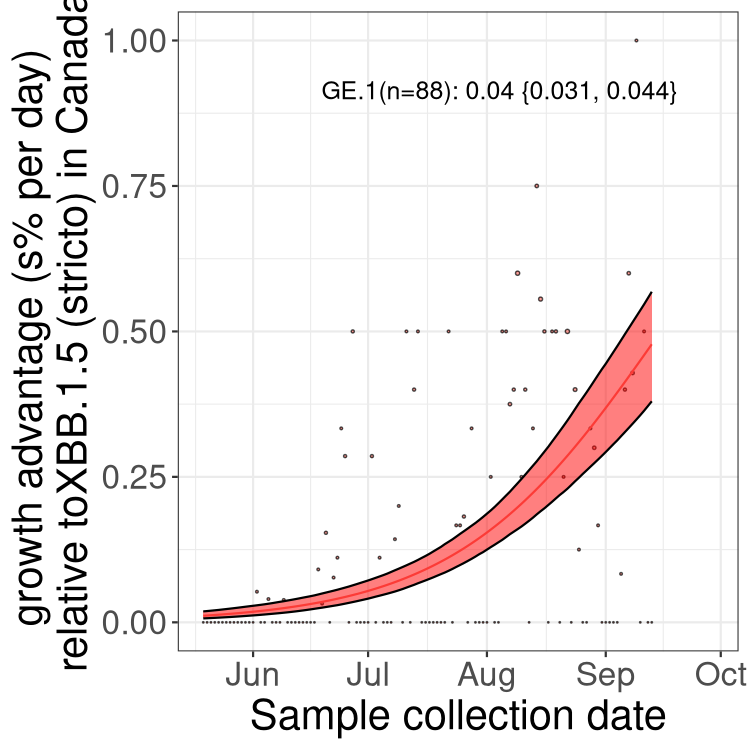
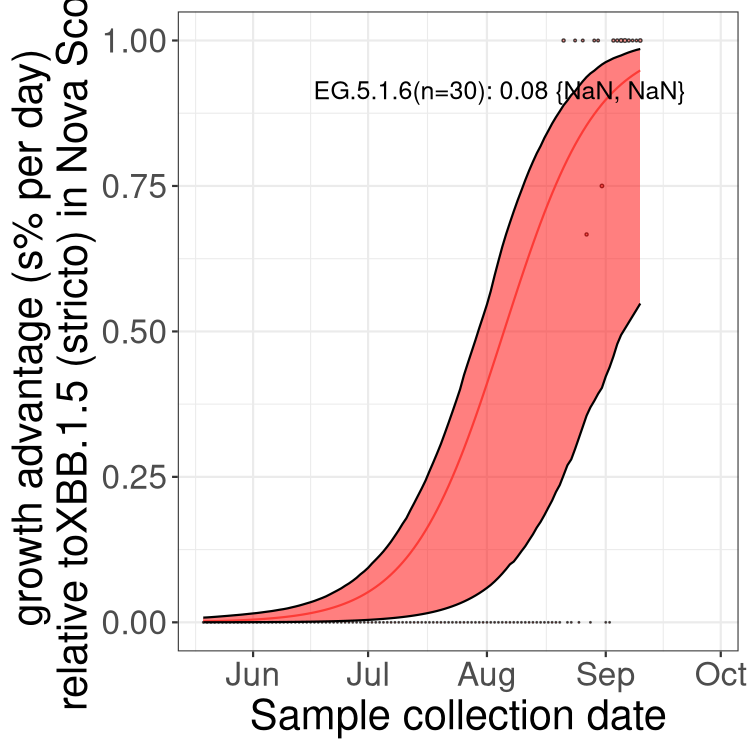
BA.2 and XBB sublineages
Here we show the trends of the various BA.2.* sublineages over time, relative to the frequency of XBB.1.5 by itself (shown for sublineages with at least 50 (Canada) or 20 (provinces) cases). Proportions shown here are only among XBB.1.5 (stricto) and the lineage illustrated. Note that these plots are not necessarily representative of trends in each province and that mixing of data from different provinces may lead to shifts in frequency that are not due to selection.
Canada
Canada



Only the three most strongly selected variants are displayed. Click here to see the rest.




































BC
British Columbia



Only the three most strongly selected variants are displayed. Click here to see the rest.




AB
Alberta



Only the three most strongly selected variants are displayed. Click here to see the rest.













SK
Saskatchawan

MB
Manitoba

ON
Ontario



Only the three most strongly selected variants are displayed. Click here to see the rest.






































QC
Quebec



Only the three most strongly selected variants are displayed. Click here to see the rest.











NS
Nova Scotia

Only the three most strongly selected variants are displayed. Click here to see the rest.




NB
New Brunswick



NL
Newfoundland and Labrador


NULL
BA.5 sublineages
Here we show the trends of the various BA.5* sublineages over time, relative to the frequency of XBB.1.5 by itself (shown for sublineages with at least 50 (Canada) or 20 (provinces) cases). Proportions shown here are only among XBB.1.5 (stricto) and the lineage illustrated. Note that these plots are not necessarily representative of trends in each province and that mixing of data from different provinces may lead to shifts in frequency that are not due to selection.
Canada
Canada

BC
British Columbia

AB
Alberta

SK
Saskatchawan

MB
Manitoba

ON
Ontario

QC
Quebec

NS
Nova Scotia

NB
New Brunswick

NL
Newfoundland and Labrador

NULL
COVID-MVP: Mutational composition of Omicron
An preview of the mutational profile of actively circulating strains and its sublineages in Canada are shown above. This preview is from https://virusmvp.org/covid-mvp.
Variants in Canada over time
This plot shows the changing composition of sequences for all Canadian data posted to the VirusSeq Portal according to Pango lineage designation (Pango version 4.2 (Viral AI)), up to 2023-09-23. Because sampling and sequencing procedures vary by region and time, this does not necessarily reflect the true composition of SARS-CoV-2 viruses in Canada over time.
Historical notes
From the beginning of the pandemic to the fall of 2021, Canadian sequences were mostly of the wildtype lineages (pre-VOCs). By the beginning of summer 2021, the VOCs Alpha and Gamma were the most sequenced lineages overall in Canada. The Delta wave grew during the summer of 2021 with sublineages AY.25 and AY.27 constituting sizeable proportions of this wave. Omicron arrived in November of 2021 and spread in three main waves, first BA.1* (early 2022), then BA.2* (spring 2022), then BA.5* (summer 2022). Current, multiple sublineages of Omicron persist, with emerging sublineages spreading, such as BQ.1.1 (a BA.5 sub-lineage).
There are two Pango lineages that have a Canadian origin and that predominately spread within Canada (with some exportations internationally): B.1.438.1 and B.1.1.176. Other lineages of historical interest in Canada:
- A.2.5.2 - an A lineage (clade 19B) that spread in Quebec, involved in several outbreaks before Delta arrived (see this post for more details: https://virological.org/t/recent-evolution-and-international-transmission-of-sars-cov-2-clade-19b-pango-a-lineages/711)
- B.1.2 - a USA lineage that spread well in Canada
- B.1.160 - an European lineages that spread well in Canada
This historical analysis is not being further updated, as we focus on more interactive data plots and the “Current situation” text above.
Canadian trees
Here we present a subsampled phylogenetic snapshot of SARS-CoV-2 genomes from Canada. The x-axis of the time tree represents the estimated number of years from today for which the root emerged. Due to the low number of XBB sequences, this estimate may not be accurate for the XBB* time tree. The x-axis of the diversity trees shows the number of mutations from the outgroup. Clicking on a node with the information tooltip shown will copy the isolate ID to your clipboard.
### metadata and trees
source("scripts/tree.r")
# load trees from files
mltree <- read.tree(paste0(params$datadir,"/aligned_nonrecombinant_sample1.rtt.nwk"))
ttree <- read.tree(paste0(params$datadir,"/aligned_nonrecombinant_sample1.timetree.nwk"))
recombTTree <- read.tree(paste0(params$datadir,"/aligned_recombinant_X_sample1.timetree.nwk"))
recombMLree <- read.tree(paste0(params$datadir,"/aligned_recombinant_X_sample1.rtt.nwk"))
#stopifnot(all(sort(mltree$tip.label) == sort(ttree$tip.label)))
dateseq <- seq(ymd('2019-12-01'), ymd('2022-12-01'), by='3 month')
# tips are labeled with [fasta name]_[lineage]_[coldate]
# extracting just the first part makes it easier to link to metadata
mltree$tip.label <- reduce.tipnames(mltree$tip.label)
ttree$tip.label <- reduce.tipnames(ttree$tip.label)
recombTTree$tip.label <- reduce.tipnames(recombTTree$tip.label)
fieldnames<- c("fasta_header_name", "province", "host_gender", "host_age_bin",
"sample_collected_by", "purpose_of_sampling",
"lineage", "pango_group","week", "GID")
# extract rows from metadata table that correspond to ttree
metasub1 <- meta[meta$fasta_header_name%in% ttree$tip.label, fieldnames]
# sort rows to match tip labels in tree
metasub1 <- metasub1[match(ttree$tip.label, metasub1$fasta_header_name), ]
#omi tree metadata
#metasub_omi <- metasub1[grepl("Omicron",metasub1$pango_group ), ]
#recomb tree metadata
mmetasub_recomb <- meta[meta$fasta_header_name%in% recombTTree$tip.label, fieldnames]
mmetasub_recomb <- mmetasub_recomb[match(recombTTree$tip.label, mmetasub_recomb$fasta_header_name), ]
#scale to number of mutations
mltree$edge.length <- mltree$edge.length*29903
mltree <- ladderize(mltree, FALSE)
recombMLree$edge.length <- recombMLree$edge.length*29903
recombMLree <- ladderize(recombMLree, FALSE)
#enforce a non zero branch length so lines can be drawn in javascript
###Time Tree
ttree$edge.length[ttree$edge.length == 0] <- 1e-4
#ttree <- ladderize(ttree, FALSE)
recombTTree$edge.length[recombTTree$edge.length == 0] <- 1e-4
#recombTTree <- ladderize(recombTTree, FALSE)
hab=unique(meta$host_age_bin)
hab=hab[order(hab)]
months=unique(meta$month)
months=as.character(months[order(months)])
weeks=unique(meta$week)
weeks=as.character(weeks[order(weeks)])
presetColors=data.frame(name=c("other",
VOCVOI$name,
hab,
months,
weeks),
color=c("#777777",
VOCVOI$color,
rev(hcl.colors(length(hab)-1, "Berlin")),"#777777",
hcl.colors(length(months), "Berlin"),
hcl.colors(length(weeks), "Berlin")
))
#suppressWarnings({
# res <- ace(metasub1$pango.group, ttree2, type="discrete", model="ER")
#})
#idx <- apply(res$lik.anc, 1, which.max)[2:nrow(res$lik.anc)] # exclude root edge
#anc <- levels(as.factor(metasub1$pango.group))[idx]
source("scripts/tree.r")
timeTreeJsonObj <- DrawTree(ttree, metasub1, "timetree", presetColors, fieldnames=fieldnames)
recombTimeTreeJsonObj <- DrawTree(recombTTree, mmetasub_recomb, "recombtimetree", presetColors, "lineage", fieldnames= fieldnames)
#diversity ML tree
diversityTreeJsonObj <- DrawTree(mltree, metasub1, "mltree", presetColors, fieldnames=fieldnames)
recombDiversityTreeJsonObj <- DrawTree(recombMLree, mmetasub_recomb, "recombmltree", presetColors, "lineage", fieldnames=fieldnames)
#write(recombDiversityTreeJsonObj, "downloads/test.json")
### omicron diversity tree
#MLtree_omi<-keep.tip(mltree, metasub_omi$fasta_header_name)
#OmicrondiversityTreeJsonObj <- DrawTree(MLtree_omi, metasub_omi, "omimltree", presetColors, fieldnames=fieldnames)